Hansen Solubility Parameters in Practice (HSPiP) e-Book Contents
(How to buy HSPiP)
Chapter 23, Noses artificial and natural (HSP for Sensors Both Artificial and Live)
Professor William Goddard’s group at
CalTech provides a good example of how HSP can be used to investigate both
artificial and natural noses. Along the way, the group is also providing new
tools for calculating HSP. We are grateful to Professor Goddard and his colleague
Dr Mario Blanco for giving us access to their data and insights.
The
artificial nose
A fruitful approach to producing an
artificial nose is to provide a sensor made up of an array of sub-sensors each
of which has a different response to an odorant molecule. Whilst the response
from any individual sub-sensor might not tell you too much about an odorant,
the “fingerprint” of responses might be sufficiently unique to tell you what
odorant is there and how much of it is present.
The “JPL Artificial Nose” works in this
way. There are seven sub-sensors, each a simple polymer: Poly(methylmethacrylate)
(PMMA), Poly(4-hydroxystyrene) (P4HS), Polyethyleneoxide (PEO), Polyethylene
(PE), Poly(ethylenevinyl acetate) (PEVA), Polysulfone, and Polycaprolactone.
The hypothesis of the Goddard group is that
the amount of interaction (and therefore sub-sensor signal) of an odorant
molecule could be predicted on the basis of its HSP distance from each polymer.
Their work is described in their paper (in collaboration with 3M) M. Belmares,
M. Blanco, W. A. Goddard III, R. B. Ross, G. Caldwell, S.-H. Chou, J. Pham, P. M. Olofson, Cristina Thomas, Hildebrand and Hansen Solubility Parameters
from Molecular Dynamics with Applications to Electronic Nose Polymer Sensors, J
Comput. Chem. 25: 1814–1826, 2004. Their analysis is based on their
calculated HSP values from molecular dynamics. In this account we use
conventional HSP for the simple reason that the paper’s values for δP and δH
are necessarily constrained by the absence of an agreed methodology for
producing these values from the MD data. We are grateful for their permission
to recast the data in our mode and we must stress our enthusiasm for the MD
approach where it is surely only a matter of time before the δP/δH problem is
solved.
The aim of the paper was to see if there is
a linear relationship between theoretical and experimental response curve. The
logic is that the sensor response depends on swelling of the polymer by the
solvent and the closer the solvent is to the HSP of the polymer the more
swelling will occur. The fit was based on 5 parameters: a pre-exponential term,
a term relating to molar volume, then a term each for the absolute differences
of the cohesive energy terms values (polymer – solvent) of D, P and H.
Equ. 1‑1 Response = P1 * Exp(-P2*MV)*Exp(P3*(δDp-δDs) + P4*(δPp-δPs) + P5*(δHp-δHs))
We decided to see what happened if we used
pure HSP instead. In that case we have just 3 parameters: the pre-exponential
term, the molar volume term and term relating to the standard HSP distance.
Equ. 1‑2 Response = P1 * Exp(-P2*MV)*Exp(P3*Sqrt(4*(δDp-δDs)2 + (δPp-δPs)2
+ (δHp-δHs)2))
The results are interesting and encouraging.
With reasonable values for the δD, δP and δH values of the polymers, the fits
were better (both in terms of slope and R2) than in the original
paper. This isn’t as good as it sounds. The paper used the computed values and produced
an honest fit. Because we had no direct knowledge of the polymers used we could
“tweak” the polymer parameters (within reasonable limits) to get a good fit. To
a certain extent we could argue (see the section on Chromatography) that this
is a good way to derive HSP for polymers, but there is rather too much
circularity in that argument.
Data from two of the polymers seem to be
illuminating:
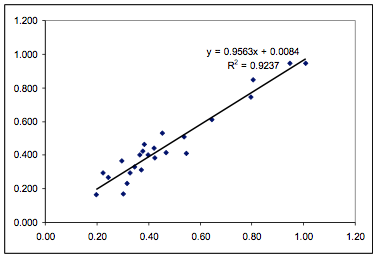
Figure 1‑1 Fit from the paper for the
Polysulfone data using 5 fitting parameters
Figure 1‑2 Fit of the Polysulfone data
using the HSP formulation and 3 parameters
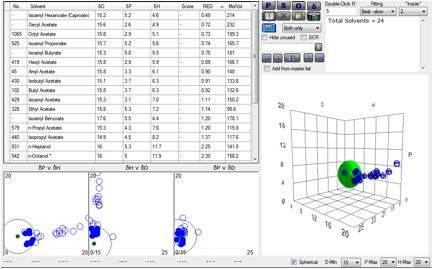
Figure 1‑3 Using file NoseChems and a Polysulfone of [15.4, 4.5, 2.8] There’s a reasonable mixture of overlapping
and non-overlapping solvents, giving an overall wide-ranging response
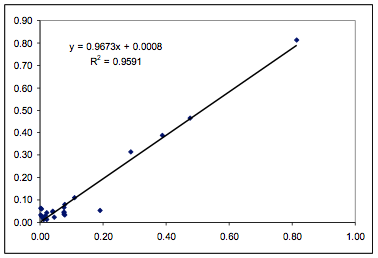
Figure 1‑4 For P4HS there is a less uniform response in the 5-parameter fit
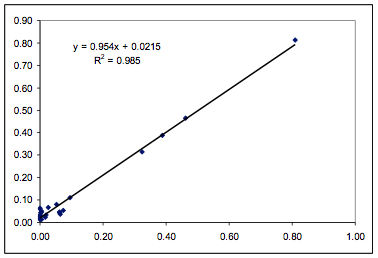
Figure 1‑5 With a slightly better, but still skewed fit from 3 parameters
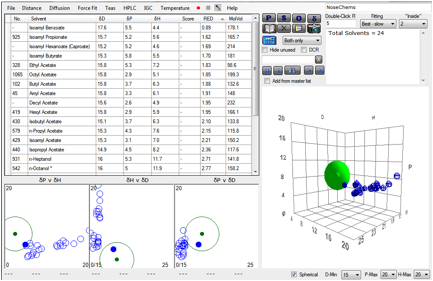
Figure 1‑6 With P4HS [18,8,2] there is very little polymer/solvent overlap, so
it’s not surprising that most responses are clustered near one end of a rather
small response curve
So at the very least the “pure” HSP approach
looks interesting. And the fact that 3 parameters suffice to fit the
experimental data from 24 solvents (using standard, un-tweaked HSP) with 7
polymers in a complex artificial nose is at the very least encouraging.
Natural
noses
We don’t want to get involved in the major
debates on how real noses manage to distinguish between so many different
aromas. But it seems reasonable to most people that unless the aroma molecule
has some affinity for a receptor site then that site won’t be able to detect it.
And as soon as the word “affinity” is mentioned, it becomes natural to ask
whether HSP could be a significant part of that affinity, and therefore a
significant predictor of smell. We say “significant” because it is unrealistic
to expect that every receptor is simply HSP generic. We are all familiar with
the fact that biological receptors can be exquisitely specific (especially when
it comes to optical isomers). So it seems reasonable that there will be
elements of specificity in nasal receptors. But what seems to be clear is that
no model based strongly on biological-style specificity has proven to be of
general utility.
So how might one show that HSP can be
insightful for understanding olfaction? The hypothesis from Blanco and Goddard
(BG) is elegantly simple. For consistency with the rest of the book (and the
eNose example above) we recast their hypothesis (with their permission) in a
slightly different formulation but the effect is the same. They used Mean Field
Theory (MFT) as their descriptor but we can think of it as HSP theory.
The
BG-HSP hypothesis
Professor Linda Buck won the Nobel prize
for identifying 47 different olfactory receptors. The BG-HSP hypothesis states
that each of these olfactory receptors is defined by a δD, δP, δH and Radius as
if it were a polymer. The response of each receptor to an odorant depends on
the HSP distance of the odorant from the receptor.
Thus the Responsejk of receptor
k to odorant j is given by
Equ. 1‑3 Responsejk = Sk * Exp(-ak*Sqrt(4*(δDk-δDj)2 + (δPk-δPj)2 + (δHk-δHj)2))
which you will recognise as being almost
identical to the eNose formula above – without the molar volume term. The
formula can be made even more familiar with one simple change:
Equ. 1‑4 Responsejk = Sk * Exp(-Sqrt(4*(δDk-δDj)2
+ (δPk-δPj)2 + (δHk-δHj)2)/Rk)
where we have replaced ak with the more familiar Radius term Rk – so that
the response has decreased by a factor of 1/e by the time the HSP distance is
equal to Rk.
The beauty of this formula is that it can
readily be tested and BG have provided the first HSP values for olfactory
receptors.
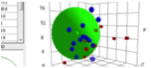
The response of the S19 receptor to 19
odorants is shown. The Sphere fit gives the HSP for S19 to be ~ [16.3, 5.5, 8.5].
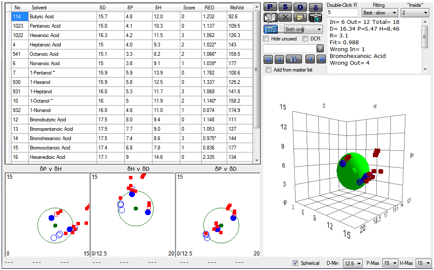
Figure 1‑7 Using file OlfactionS19
The S83 receptor gives: [16.4, 4.7, 8.4]:
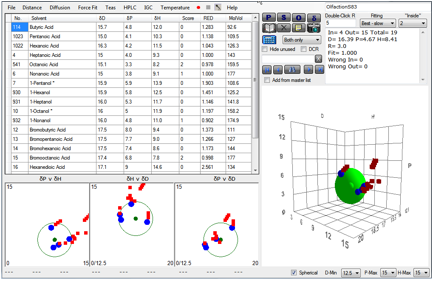
Figure 1‑8 Using file OlfactionS83
The process can be repeated for the other
receptors. Note that the S83 receptor is more specific (R=3) than the S19
receptor (R=4).
When you try these examples out for
yourself you will quickly find that we have shown the best possible
interpretation of these data. The data sets are too small to provide good fits
and it would be a massive task to take on such a vast project.
But as the original BG paper shows, the
idea is, at the very least, a very fertile one. If olfaction is, as they
guesstimate, 65% HSP and 35% specific receptor, then there will be plenty of
noise in the data, but the HSP signal should shine through if the hypothesis is
correct. And of course, life can be more complicated. Perhaps (and BG have
evidence for this) some receptors have two HSP sites. A single Sphere fit would
not do a good job so a more sophisticated multi-Sphere calculator would be
required.
The reason the BG-HSP hypothesis is so
important is because if it were shown to be true it would not be
yet-another-correlation but a deep insight into olfactory receptors. Given that
HSP are calculable ab-initio from molecular dynamics, and given that HSP
represent fundamental thermodynamics, then olfaction (to, say, 65%) would
become calculable from first principles.
If by the time you read this book the
ongoing research has confirmed BG-HSP then we will be pleased that we spotted
the significance of the BG research before it had reached maturity. If it has
been disconfirmed then we’re pleased in another sense. For HSP to be good
science it has to withstand the harsh standard of disconfirmation. If it has
proven to have failed on such a big task as olfaction it at least had the merit
of offering a clear prediction which could
be refuted, and that’s one of the hallmarks of good science.
The
Atlas of Odor Character Profiles
The Atlas, by Andrew Dravnieks, published
by ASTM, ISDN 0-8031-0456-1, is a book of tables. For 144 odour chemicals plus
a few more odour mixtures it lists to what extent a panel of skilled testers
would say that each chemical smelled like X, where X was a list of 146
different odour sensations such as Fruity, Almond, Molasses, Yeasty, Incense,
Kerosene, Sweaty, Heavy – to take a random cross-section through that list
of sensations.
In the spirit of making refutable
predictions, it seemed a good idea to assemble the HSP of all 144 chemicals
then see how these fitted to the odour profiles. Of the 144 chemicals, many
were in the standard Hansen table, but most were not. Thanks to the generosity
of SC Johnson, a list of HSPs of 288 odour chemicals (prepared using
calculation/estimation only by Charles Hansen) was made publicly available.
This still left quite a few of the 144 without HSP so Abbott used the DIY-HSP
tools from HSPiP to estimate the remaining ones. Both the SC Johnson list and
the 144 list are made available as a contribution to further research on odours
and fragrances. The 144 list includes CAS Numbers and Smiles notation to help
you make sure which chemical is being referred to - naming of odorants is
rather uncertain. Note that there are some minor errors in the Atlas. Where
possible the table contains revisions to these errors.
Matching chemicals to the different
sensations was made possible thanks to further tables in the Atlas. These
listed the 5 highest-scoring chemicals for each sensation. The “minimum HSP
sphere” that enclosed these 5 chemicals was then used as an indication of the
hypothetical HSP centre/radius for each of these sensations.
Of course there are many problems with this
procedure. First, some of the top 5 chemicals were from the mixtures which have
not been included in the HSP list. Second, some of the “top 5” have such low
scores as to make it seem unlikely that these sensations really do have
well-defined chemical correlations. Third, the choice of 5 is rather arbitrary.
If the size of responses (where a large number means a strong response) go (43,
41, 38, 34, 11) should that 5th chemical (clearly much less relevant
than the other four) be included? Or if the responses went (43, 41, 38, 34, 30,
29, 28, 11) shouldn’t we include the top 7 chemicals?
But we have to start somewhere. We’re only
trying to explore some basic hypotheses. Others can feel free to refine the
process if it seems to be worthwhile.
Although it was a lot of work, the “easy”
part of the process was to identify the HSP centre/radius for each relevant
sensation. The raw data are provided for you to save you the tedium of creating
it for yourself. There were 70 aromas with meaningful high scores for which the
centres of the minimum spheres were calculated.
Here is a screenshot from HSPiP showing 71
aromas in HSP space:
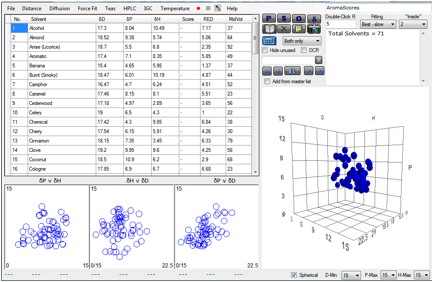
Figure 1‑9 Using file AromaScores
The hard part is working out whether the
data mean anything. The simplest case would be that each sensation had a unique
sensor which had a unique HSP for optimal binding. It’s obvious that aromas
cannot work this way. Some of the sensations must be complex mixes of different
sub-sensations. And the most optimistic HSP case would be that HSP
compatibility is necessary but not sufficient – there must be a good HSP
match for a molecule to be happy in the sensor area, but there must be other
molecule-specific attributes for the aroma to register with the sensor.
An alternative would be to follow the
process of A.M. Mamlouka, C. Chee-Ruiter, U.G. Hofmann, J.M. Bower, Quantifying olfactory perception: mapping
olfactory perception space by using multidimensional scaling and self-organizing
maps, Neurocomputing 52–54, 2003, 591 – 597. For those who are
familiar with multidimensional scaling and self-organizing maps the data from
our explorations are provided. However, the best that Abbott could do was to
prepare a spreadsheet with a 71x71 matrix that calculated the HSP distance
between each of the aromas. It was then possible to sort each column to see if
the ordering of the aromas made sense. For example, if the target aroma was
“bananas” which is arguably a pleasant aroma, other pleasant aromas might be
expected to be close by and disgusting odours would be far away.
There was, unfortunately, no compelling
evidence for this happy outcome. Here is a small section of the matrix ordered
by distance between Banana and other aromas. Some of the fruity odours are
gratifyingly close to Banana, but Urine and Rancid are also fairly close and
they are not normally associated with the aroma of Banana.
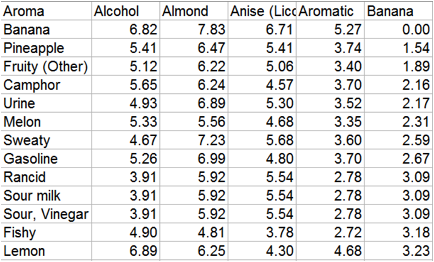
Figure 1‑10 A portion of the matrix ordered by the HSP distance between Banana
and the other 70 odours.
Hansen published a paper in 1997 (Hansen,
C.M., Aromastoffers
Opløselighedsparametre (in Danish), Solubility Parameters for Aromas and Scents. Plus Process, 11,
16-17,1997) which anticipated many of these ideas. Readers may or may not like
to know that it is possible to cover the smell of skatole (faeces) with
suitably chosen (i.e. a good HSP match) aromas from hamburgers or bacon.
3rd
Edition update
With the benefit of hindsight, some of the
ideas above do look naive. But it still seems to us that the world of
fragrances is missing a trick by not taking HSP into account.
At the very least, the packaging industry
could get a lot of benefit from the ideas of HSP and diffusion. If there is a
good HSP match of a key fragrance/flavour component, say, Cinnamon
(cinnamaldehyde) with a packaging polymer (such as poly lactic acid, PLA) then
it’s highly likely that the polymer will be a poor barrier for it.
Similarly, if a fragrance component (or,
more likely, fragrance formulation) has a good match for the HSP of skin then
it’s more likely to penetrate the skin and (most likely) be lost as an odour.
And clearly if a fragrance/flavour
component is to be delivered within some polymeric system (e.g. scratchable
spheres) classic HSP calculations will help ensure a balance of good
compatibility for creating the system and poor compatibility to ensure that the
fragrance remains locked in to the system till required.
Because Sigma Aldrich have provided a
de-facto standard reference for aromas, with helpful designation of the
different types of smell, we are putting into the public domain an HSPiP
version of the Sigma Aldrich Flavors & Fragrances catalogue. This is a
somewhat error-prone undertaking as anyone who has ever handled complex
datasets will know from their own experience. The catalogue doesn’t always
provide CAS numbers and it doesn’t supply the Smiles. So at times we used the
also excellent GoodScentsCompany website and you might find some alternative
names for the same compound. We also had to decide what to include. Although we
could have included the “W” numbers from the catalogue, there are so many
variants of essentially the same compound that we decided it would not be helpful.
Similarly, we could have provided the aroma class, but that would have created
much duplication. From the name and/or the CAS number you should be able to
identify most of the chemicals in the catalogue and therefore their aroma
class. Finally, of course, we could not include those aromas that are mixtures
and sometimes even the wonderful ChemSpider could not help us identify the
right Smiles for a given compound. Nevertheless, you now have access to the HSP
of over 800 aroma chemicals which you can then cross-reference with the Sigma
Aldrich catalogue for your own purposes.
When you load it into HSPiP you will see
the usual HSP data. But the horizontal scroll bar will allow you to scan across
for the Smiles, CAS number etc. To search within the data you can try using a
name (probably a truncated version as naming is so variable) or a CAS number.
One way of exploring whether HSP have any
relation to aromas is via a Self Organising Map (SOM). Hiroshi has enjoyed
playing with this concept and the following section shows the sort of
exploration that can be done. It’s included as an indication of what might be
done if someone wanted to throw some serious resource at the issue.
10
Fragrant Flowers
A Japanese website http://www001.upp.so-net.ne.jp/iromizu/hana_kaori_for_so-net.html
lists the key ingredients of 10 flowers:
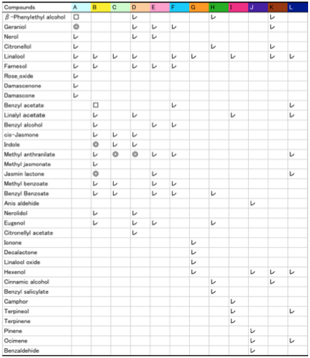
From the HSP of these molecules, and the
different fragrances, an SOM can be constructed on a 40x40 matrix:
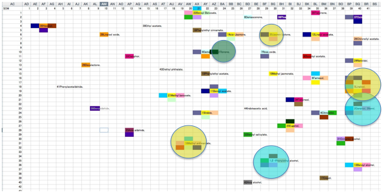
From this partition it’s possible to ask
many questions. A typical one is “can we distinguish some key distinct notes
from this?” And here’s one answer. 5 distinct areas stand out: Rose,
Orange-colour olive, Lilac, Carnation and Jasmine stand out.
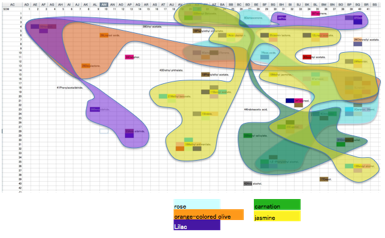
On the other hand, Ixora, Narcissus,
Jasmine are rather similar in SOM space:
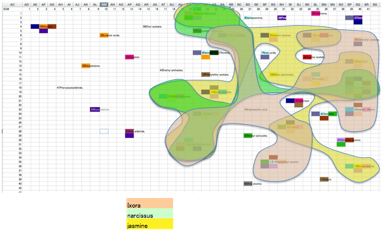
As we’re not trained in fragrances we can’t
comment on the significance of these plots. But it represents another way of
looking at how fragrances and solubilities may be related. If they show no
relationship, that’s useful to know. If there are such relationships (and the
physiology suggests that there should be) then HSP provide an opportunity for
data mining in this fascinating area.
E-Book contents | HSP User's Forum