HSP Application note #61
Casting Solvents for membrane2010.11.19
HSPiP Team Senior Developer, Dr. Hiroshi Yamamoto
I got interesting paper.
Poly(vinylidene fluoride-co-hexafluoropropene) (PVDF-HFP) membranes for ethyl acetate removal from water
Xiuzhi Tian ∗, Xue Jiang
School of Textiles & Clothing, Key Laboratory of Eco-Textiles, Ministry of Education, Jiangnan University, Wuxi 214122, China
Journal of Hazardous Materials 153 (2008) 128–135
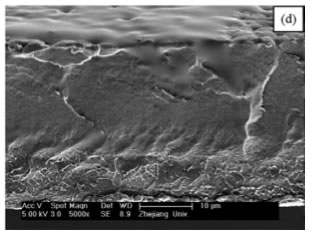
SEM pictures of PVDF-HFP membranes’ cross-sections prepared under different conditions.
DMAc, vacuum, 60 ◦ C.
They cast PVDF-HFP polymer with several conditions.
In this paper, author use Hansen solubility parameters (HSP) of PVdF-HFP [17.2, 12.5, 8.2].
But reference paper, it is not PVdF-HFP but pure PVdF.
And the HSP value is [17.2, 12.5, 9.2] in original paper.
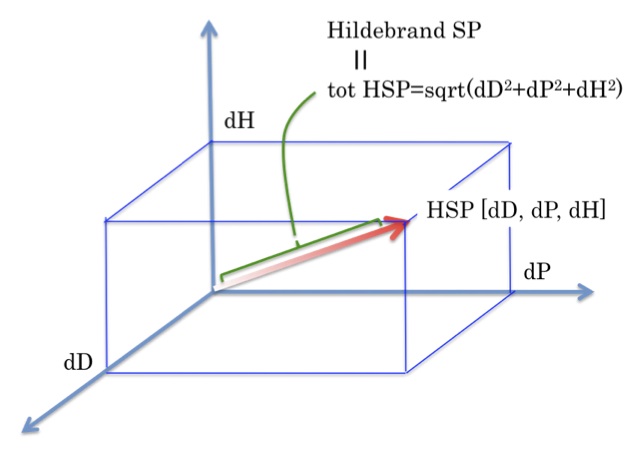
Hansen Solubility Parameters (HSP)Hansen Solubility Parameters(HSP) were developed by Charles M. Hansen as a way of predicting if one material will dissolve in another and form a solution. They are based on the idea that "like dissolves like" where one molecule is defined as being 'like' another if it bonds to itself in a similar way.
What can perhaps be surprising is that one can assign HSP to so many different things. Gases like carbon dioxide, solids like carbon-60, sugar, and biological materials like human skin, depot fat, DNA, and even some proteins all have HSP. The list can be continued with drugs, polymers, plasticizers, and in fact any organic material and even many inorganic materials like salts. The only requirement for an experimental confirmation is that the material must behave differently in a sufficient number of test solvents upon contact. Pirika JAVA Demo Applet calculate HSP. HSPLight is available here. |
The distance calculation is wrong with this paper.
δiP = [(δpi − δpP)2 + (δdi − δdP)2 + (δhi − δhP)2]1/2 (7)
It need 4* before δd term.
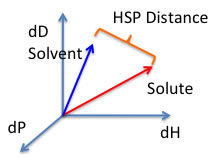
HSP DistanceTo calculate the distance (Ra) between Hansen parameters in Hansen space the following formula is used:
HSP distance(Ra)={4*(dD1-dD2)2 + (dP1-dP2)2 +(dH1-dH2)2 }0.5 |
Anyway, I try to use HSP for “Choice of Solvents for membrane preparation”.
Drag=Rotate, Drag+Shift=Larger/Smaller, Drag+Alt or Command(Window key)=Translate.
If you are using HTML5 enable browser such as Chrome, Safari or FireFox (IE9 is out of support), you will see the Canvas. If you pick solvent, solvent name will appear.
As I wrote at PVdF binder resin, this polymer has double natures.
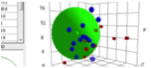
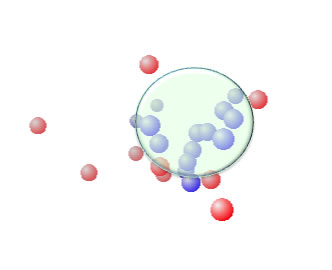
1st Large Green Sphere, [19.1, 15.6, 10.2] Radius 8.45
2nd Small Green Sphere, [17.5, 6.3, 9.0] Radius 4.63
| Hcode | Name | dD | dP | dH | DistSP-A | Dis/RA | DistSP-B | Dist/RB |
| 417 | hexane | 14.9 | 0 | 0 | 20.44 | 2.42 | 12.15 | 2.63 |
| 181 | cyclohexane | 16.8 | 0 | 0.2 | 19.09 | 2.26 | 10.91 | 2.36 |
| 456 | methyl alcohol | 14.7 | 12.3 | 22.3 | 15.32 | 1.81 | 15.63 | 3.38 |
| 325 | ethyl alcohol | 15.8 | 8.8 | 19.4 | 13.21 | 1.56 | 11.22 | 2.42 |
| 569 | propyl alcohol | 16 | 6.8 | 17.4 | 12.95 | 1.53 | 8.93 | 1.93 |
| 92 | butanol | 16 | 5.7 | 15.8 | 12.95 | 1.53 | 7.46 | 1.61 |
| 182 | cyclohexanol | 17.4 | 4.1 | 13.5 | 12.44 | 1.47 | 5.01 | 1.08 |
| 46 | aniline | 20.1 | 5.8 | 11.2 | 10.05 | 1.19 | 5.67 | 1.22 |
| 11 | acetophenone | 18.8 | 10 | 4 | 8.38 | 0.99 | 6.74 | 1.46 |
| 363 | ethylene carbonate | 18 | 21.7 | 5.1 | 8.25 | 0.98 | 15.92 | 3.44 |
| 584 | Propylene Carbonate | 20 | 18 | 4.1 | 6.80 | 0.80 | 13.63 | 2.94 |
| 115 | gamma-butyrolactone | 18 | 16.6 | 7.4 | 3.70 | 0.44 | 10.47 | 2.26 |
| 617 | tetrahydrofuran | 16.8 | 5.7 | 5.7 | 11.81 | 1.40 | 3.63 | 0.79 |
| 285 | N,N'-dimethylacetamide | 16.8 | 11.5 | 10.2 | 6.16 | 0.73 | 5.52 | 1.19 |
| 521 | N-methyl-2-pyrrolidone | 18 | 12.3 | 7.2 | 4.97 | 0.59 | 6.34 | 1.37 |
| 303 | dimethyl sulfoxide | 18.4 | 16.4 | 10.2 | 1.61 | 0.19 | 10.33 | 2.23 |
| 624 | Tetramethylurea | 16.7 | 8.2 | 11 | 8.86 | 1.05 | 3.19 | 0.69 |
| 416 | hexamethyl phosphoramide | 18.5 | 11.6 | 8.7 | 4.44 | 0.53 | 5.67 | 1.23 |
| 659 | triethyl phosphate | 16.7 | 11.4 | 9.2 | 6.46 | 0.76 | 5.35 | 1.16 |
| 671 | trimethyl phosphate | 15.7 | 10.5 | 10.2 | 8.50 | 1.01 | 5.66 | 1.22 |
| 297 | N,N'-dimethylformamide | 17.4 | 13.7 | 11.3 | 4.05 | 0.48 | 7.75 | 1.67 |
| 183 | cyclohexanone | 17.8 | 8.4 | 5.1 | 9.20 | 1.09 | 4.47 | 0.97 |
If JAVA 3D is already installed in your machine, please try 3D view of this Sphere.
Hansen SphereTo determine if the parameters of two molecules (usually a solvent and a polymer) are within range a value called interaction radius (R0) is given to the substance being dissolved. This value determines the radius of the sphere in Hansen space and its center is the three Hansen parameters.
From version 3.1.X, Double Spheres function is available. Pirika provide JAVA 3D Demo Applet to browse the Sphere(s). |
I selected several solvents and calculate (solvent-polymer) distance.
If the Distance is within sphere radius, that solvent is good solvent. So I append 2 column Ra=Distance/Radius.
If Ra<1.0, then that is good solvent.
There is no solvent that dissolve both parts of polymer.
Maybe, N,N’-Dimethylacetamide (DMAc) is the most good balanced solvent.
The second example is,
MIT OpenCourseWare
2.500 Desalination and Water Purification
Amphiphilic Graft Copolymers for Nanofiltration Membranes with Tunable Pore Size
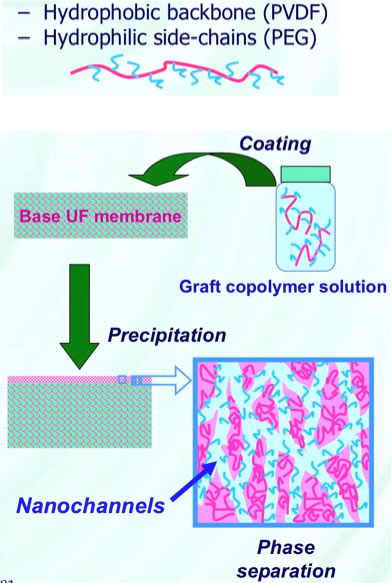
How can we use HSP designing solvents for this purpose?
At first, we need to determine HSP of PEG.
I got Solubility data from Polymer Handbook.
| Hcode | Name | dD | dP | dH | Vol | Score |
| 417 | hexane | 14.9 | 0 | 0 | 131.4 | 0 |
| 7 | acetone | 15.5 | 10.4 | 7 | 73.8 | 0 |
| 481 | methyl ethyl ketone | 16 | 9 | 5.1 | 90.2 | 0 |
| 255 | diethyl ether | 15.49 | 2.9 | 4.6 | 104.7 | 0 |
| 522 | methyl tert-butyl ether | 14.8 | 4.3 | 5 | 119.8 | 0 |
| 306 | 1,4-dioxane | 17.5 | 1.8 | 9 | 85.7 | 0 |
| 52 | benzene | 18.4 | 0 | 2 | 89.5 | 1 |
| 637 | toluene | 18 | 1.4 | 2 | 106.6 | 1 |
| 456 | methyl alcohol | 14.7 | 12.3 | 22.3 | 40.6 | 1 |
| 10 | acetonitrile | 15.3 | 18 | 6.1 | 52.9 | 1 |
| 524 | dichloromethane | 17 | 7.3 | 7.1 | 64.4 | 1 |
| 156 | chloroform | 17.8 | 3.1 | 5.7 | 80.5 | 1 |
| 122 | carbon tetrachloride | 17.8 | 0 | 0.6 | 97.1 | 1 |
| 183 | cyclohexanone | 17.8 | 8.4 | 5.1 | 104.2 | 1 |
| 202 | diethylene glycol dimethyl ether | 15.7 | 6.1 | 6.5 | 142.9 | 1 |
| 328 | ethyl acetate | 15.8 | 5.3 | 7.2 | 98.6 | 1 |
| 617 | tetrahydrofuran | 16.8 | 5.7 | 5.7 | 81.9 | 1 |
| 285 | N,N'-dimethylacetamide | 16.8 | 11.5 | 10.2 | 93 | 1 |
| 297 | N,N'-dimethylformamide | 17.4 | 13.7 | 11.3 | 77.4 | 1 |
| 303 | dimethyl sulfoxide | 18.4 | 16.4 | 10.2 | 71.3 | 1 |
| 598 | pyridine | 19 | 8.8 | 5.9 | 80.9 | 1 |
| 1016 | ethylacetoacetate | 16.5 | 7.3 | 8.3 | 127.3 | 1 |
And calculate with Sphere program.
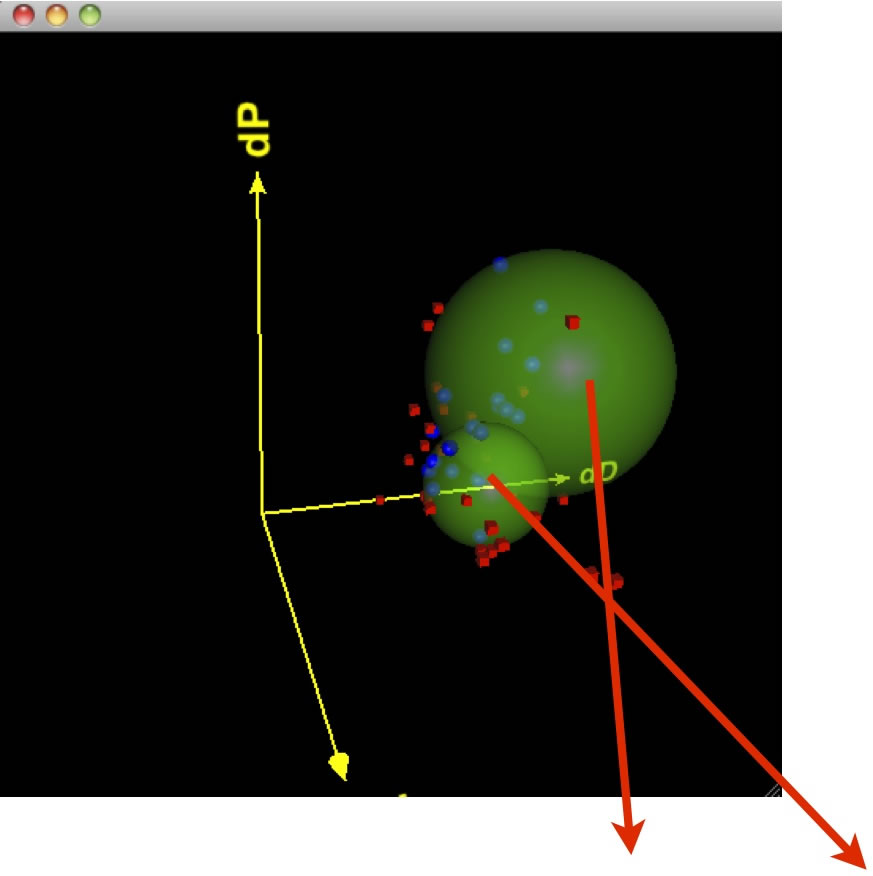
The result is [20.0, 11.2, 2.3] Radius 12.2
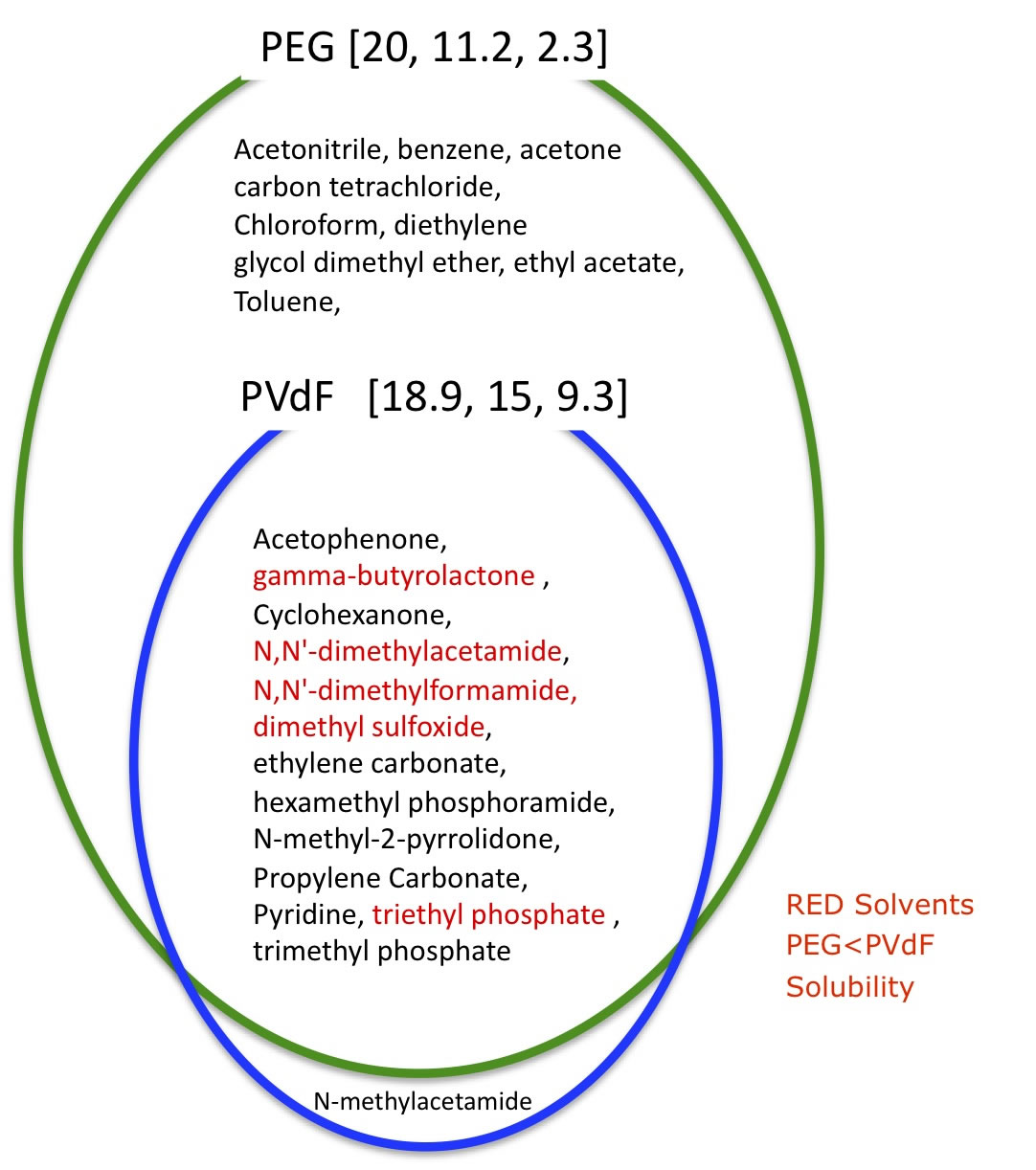
Only N-methylacetamide is good solvents for PVdF, and bad solvent for PEG.
So, this solvent might make phase separation most effectively.
(At least, you would better use RED SOLVENTS )
Drag=Rotate, Drag+Shift=Larger/Smaller, Drag+Alt or Command(Window key)=Translate.
If you are using HTML5 enable browser such as Chrome, Safari or FireFox (IE9 is out of support), you will see the Canvas. If you pick solvent, solvent name will appear. The Green Large sphere means PEG, the Cyan Sphere means PVdF.
Intrinsic Viscosity of polymer solution.