I calculated the Hansen Solubility Parameters (HSP) of the anti-cancer drug. When plotted in Hansen space with the usual solvents, there are not many solvents near the anti-cancer drug.
Now, let’s plot it with nucleosides.
The large translucent spheres are drawn as radius 5 at the Hansen Solubility Parameters (HSP) of the nucleosides. Each color is as follows.
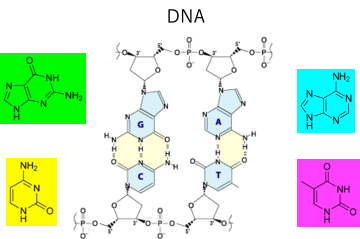
In DNA, A-T and G-C make hydrogen bonds to form a double-helix structure.
| nucleoside | Smiles | Color |
| Guanine | O=C2/N=C(Nc1ncnc12)N | Green |
| Adenine | NC1=NC=NC2=C1N=CN2 | Cyan |
| cytosine | NC1=NC(NC=C1)=O | Yellow |
| thymine | Cc1c[nH]c(=O)[nH]c1=O | Magenta |
It is interesting to note that anticancer drugs are placed around the border of these nucleoside spheres.
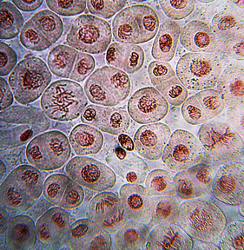
This DNA is also called chromosomes because it is easily stained and can be seen when cells are stained.
This is carminic acid dissolved in acetic acid.
Normally, it should be chelated with iron ion or aluminum ion, but let’s just calculate the HSP of the skeleton.
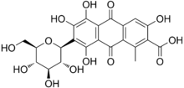
| dye | smiles |
| carminic acid | O=C(O)c2c(c3C(=O)c1c(O)c(c(O)c(O)c1C(=O)c3cc2O)[C@H]4O[C@@H]([C@@H](O)[C@H](O)[C@H]4O)CO)C |
| No Sugar | Cc2c(C(=O)O)c(O)cc3c(=O)c1c(O)c(O)cc(O)c1c(=O)c23 |
Once you have that, try to calculate the HSP distance to the nucleosides. Which nucleoside is the closest to the dye?
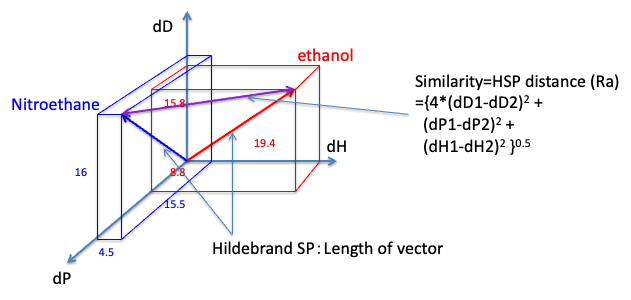
HSPiP is an indispensable software to study such solubility.
