If you have data and HSPiP, you can make a prediction system in 30 minutes.You can get various data by searching Draize Score or JaCVAM.
I downloaded the following literature from the Internet as a test.
Vitrigel-Eye Irritancy Test Method Using HCE-T Cells
toxicological sciences 135(2), 347–355 2013
The data should be compiled in this way.
The structural formula of the compound Smiles and the CAS# are brought from PubChem.
This is probably the most time consuming part.
Then, decide how to handle the HSP Score.
For example,
GHS=1 Score=1
GHS=2A Score=2
GHS=2B Score=3
GHS=NC Score=4
When ready, run the data through a converter to convert it to the hsdx file format and save it.
Read from HSPiP.
Since the Score system is a 1-6 system, choose from which level you want to be inside the Hansen’s dissolution sphere. (In this case, Inside is set to 3.) After that, just press the Calculate button and you will get the result instantly.
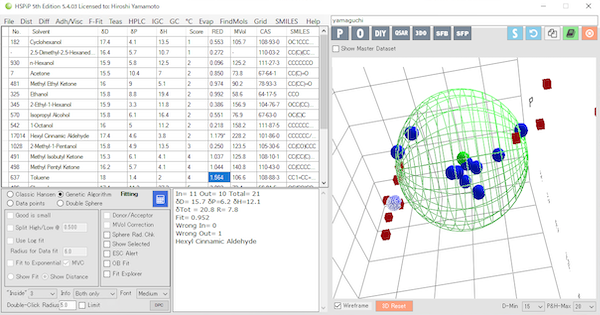
Those with GHS of 1, 2A, and 2B (stimulants) are placed inside the large green Hansen dissolution sphere, and those with GHS of NC are placed outside the sphere.
The center of the large green Hansen soluble sphere is then [δD,δP,δH]=[15.7,6.2,12.1] and the radius is 7.8.
Now a system has been created to determine whether any given compound is GHS 1, 2A, 2B (irritant) or GHS NC.
When the Hansen solubility parameter of a new compound is calculated and plotted in Hansen space, we can determine if it is an irritant if it is inside the large green Hansen solubility sphere and not an irritant if it is outside.
RED= HSP Distance / Radius can be calculated and may be done in Excel.
GHS of 1, 2A, and 2B are made Inside, so the Draize Score of 13 or higher is inside the sphere. The exception is Hexyl cinnamic aldehyde, which has a GHS of 2A and is irritating, but has a RED of 1 or higher.
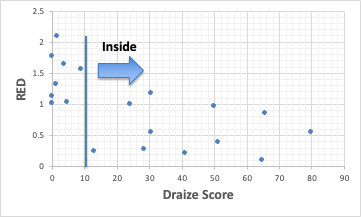
Score may be treated as a real number.
The QSAR function of HSPiP can also be used.
Whichever way you do it, you can probably build a forecasting system in 10 minutes.
It is very easy to do, so it’s great for free research during summer vacation.
Let’s try to do it by yourself at least up to this point.
Those who do it for work need to do it a little better.
This does not take into account the size of the molecule.
Naturally, large molecules are often not irritants.
And in fact, in the case of this problem, the model equation could not be created with the QSAR functionality to the degree that is available in HSPiP.
A new modeling tool from pirika, MIRAI-G, must be used.
In this case, the compound that deviates significantly from the QSAR formula is 2-Methyl-1-Pentanol.
Since the Draize Score of n-Hexanol is 64.8, it seems that the Draize Score of 13 for 2-Methyl-1-Pentanol is recognized as too small.
Hexyl cinnamic aldehyde is calculated to be 27.5, so it rides almost on a straight line; the size of the molecule of hexyl cinnamic aldehyde is the second largest of the compounds used. It is considered to be a great stimulant with a solubility of HSP that further compensates for its size.
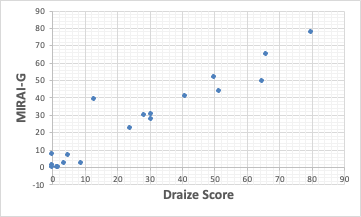
Compounds without Draize Score scores were predicted as follows.
10.7 for Methyl pentyl ketone
5.1 for Gluconolactone
Since both are below 13, they are correctly determined to be non-irritant.
The calculations for MIRAI-G take a little longer, but the nonlinearity and item interaction are taken into account. In the meantime, I will brush up the usability of the program a little more at the pirika study group and other meetings. In the future, I am thinking of offering the program as a set with a CLI license.
