Hansen Solubility Parameters in Practice (HSPiP) e-Book Contents
(How to buy HSPiP)
Chapter 4, The Grid (A different route to the Sphere)
For more than 40 years the Sphere technique
has been used with great success. But sometimes you don’t quite have the
solvent you need so you add one or two pseudo-solvents created via the mixing
rule. So if you don’t want to use dimethyl formamide in a test you can create
something close (using the Solvent Optimizer) with, say, a 60:40 mix of DMSO
and THFA. This technique has been used numerous times.
But why stop there? Why not make a whole
Sphere test from mixed solvents? If, for example, 3 pairs of solvents covered
the range of interest then instead of finding 20-30 different solvents, you can
stock just 6 of them, and if you have a robot it’s rather efficient to make up
the set of possible blends from the 3 pairs of those 6 solvents.
Although others may have done this before,
the Brabec group at U. Erlangen have shown how powerful the technique can be in
a paper that tried to narrow down the HSPs of materials used in organic
photovoltaic systems: Determination of the P3HT:PCBM solubility
parameters via a binary solvent gradient method: Impact of solubility on the photovoltaic
performance, Florian Machui, Stefan Langner, Xiangdong Zhu, Steven Abbott,
Christoph J.Brabec, Solar Energy Materials & Solar Cells 100 (2012) 138–146.
The technique seemed so powerful that for
HSPiP it was termed the Grid method (because it assembles a grid of points
throughout the 3D space). The initial version of the Grid was highly experimental
but rapidly proved so popular that it has been expanded, with the ability to
create a Grid automatically from a list of chosen solvents.
An example using the Default set is shown:
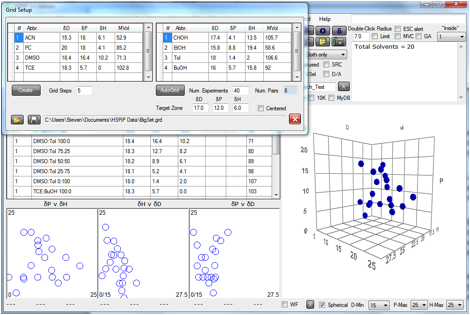
Figure 0‑1 A Grid based on the Default set
In this example 4 pairs of solvents are
chosen: Acetonitrile:Cyclohexanol; Propylene Carbonate:Ethanol; DMSO:Toluene;
Tetrachloroethylene:Butanol. Abbreviations are used because, as can be seen
from the main form, the individual points are made up of the step-wise
variation of the pairs and their full names would over-fill the space for the
names.
This particular grid is chosen because it
rather effectively fills a large part of HSP space. But of course it is not
perfect. There are some significant holes in that space. But the point of the
Grid is that it is entirely flexible. You can fill those holes by changing the
pairs or you can add a few specific single solvents. It is your choice.
This sort of Grid is general purpose. An
alternative, which is equally powerful (and is the choice in the Erlangen
paper) is to start with a single good solvent and explore the Sphere in 3
directions.
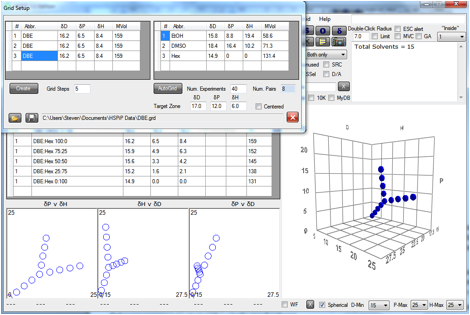
Figure 0‑2 A Grid using a single solvent (hopefully) at the centre of the
Sphere
Here the idea is that the material of
interest is soluble in DBE and by scanning out in 3 different directions the
radius and (probably) the centre can be defined more precisely than tends to
happen with the usual assortment of solvents.
One user loved the idea but wanted to be
able to create Grids automatically from a chosen set of solvents which happen
to be the only ones usable (for cost, tox, volatility reasons) in their area.
So the AutoGrid method takes a selected list of solvents (from the Solvent
Optimizer) and a Target Zone of interest and attempts to create a suitable
Grid. Finding a perfect algorithm for this has proven difficult, but it seems
that the choice of solvents is even more important than the choice of
algorithm. If, for example, Hexane:MEK turns out to be a good scan line within
the Grid, then Heptane:MEK would also be a good scan line. If the solvent list
contained both Hexane and Heptane then the chances are that both lines would be
included in the Grid (because they are both “good”), adding nothing whatsoever
to the quality of the resulting data. Although in principle a smart algorithm
can remove such pseudo-duplicates, given that humans are smarter than computers
it’s a good idea just to include heptane as it’s usually rated less toxic than
hexane.
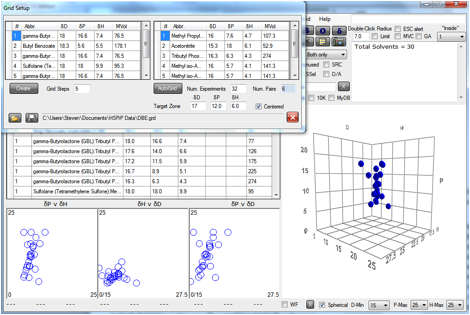
Figure 0‑3 An AutoGrid aimed at the [17, 12, 6] zone
This particular AutoGrid is limited to 32
experiments, so at 5 steps/scan-line, that means 6 pairs of solvents. The
“Centered” option was chosen to force the lines to cross the sphere as much as
possible. No doubt a human could come up with a better Grid from the given set
of solvents, but it would take much longer and at least the automated version
offers a starting point for human intervention.
A question that is often asked is: “How
good are mixed solvents compared to the same HSP from a single solvent?” The
answer from experiments carried out over 40 years is “In HSP terms they work
very well, but in kinetic terms, and in situations where entropic effects are
important they act as if they were a solvent with a larger MVol”. So if in the
example at the start DMF was a fast solvent for polymers, the DMSO:THFA mix
would be rather slower. And because entropic effects are important in polymer
solubility, the mix would not give as high a solubility as the single solvent.
This means that the Sphere radius from
mixed solvents will be rather smaller than if it had been measured with single
solvents. For most users, the ease and convenience of the Grid and the chance
of a more accurate Sphere centre seem to outweigh the issue of a potentially
smaller radius.
E-Book contents | HSP User's Forum